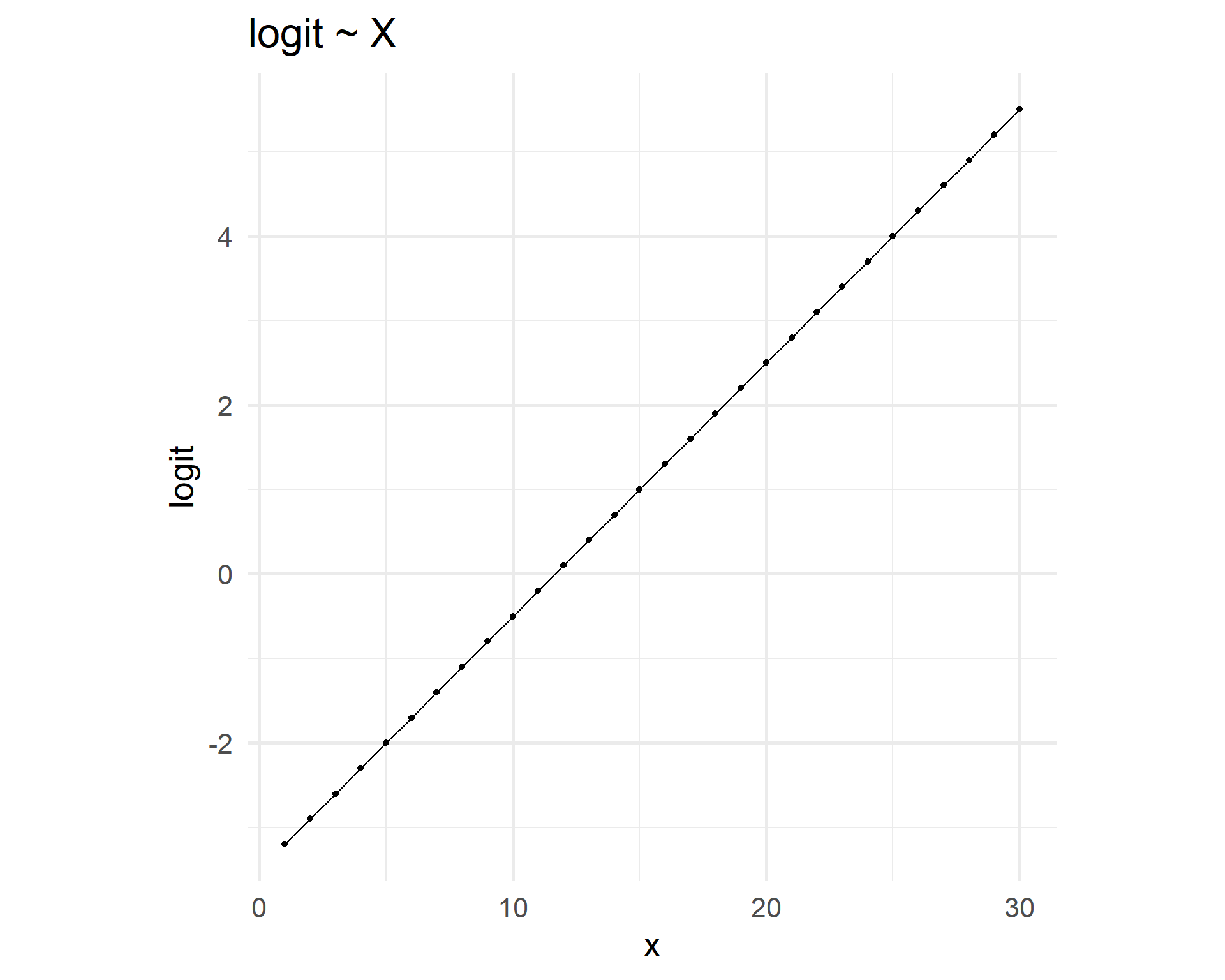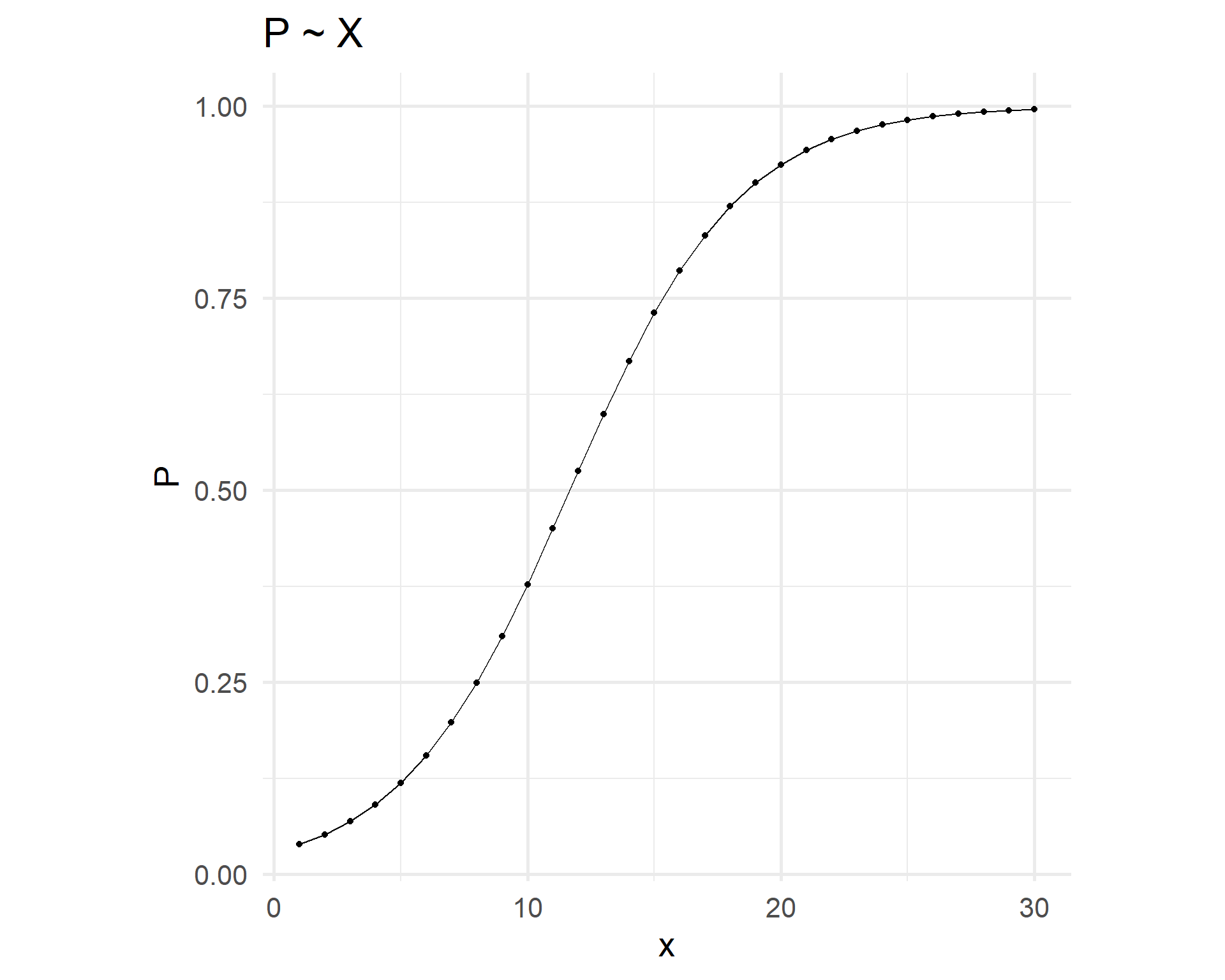set.seed(2022)
gonna <- tibble(variant = rep("gonna", 50), comp_length = rnorm(50, 6, 0.8),
register = sample(c("formal", "informal"), 50, replace = TRUE, prob = c(0.1, 0.9)))
going_to <- tibble(variant = rep("going_to", 50), comp_length = rnorm(50, 3, 1.2),
register = sample(c("formal", "informal"), 50, replace = TRUE, prob = c(0.7, 0.3)))
gt <- bind_rows(gonna, going_to) %>%
mutate(
comp_length = if_else(comp_length > 1, comp_length, 1),
variant = fct_relevel(variant, "gonna"),
variant_num = as.numeric(variant)-1,
register = fct_relevel(register, "informal"),
source = factor(sample(letters, n(), replace = TRUE))
)
m1 <- glm(variant ~ comp_length, data = gt, family = binomial(logit))
gt$fit1 <- m1$fitted.values
m2 <- glm(variant ~ comp_length + register, data = gt, family = binomial(logit))
gt$fit2 <- m2$fitted.values
gonna_plot <- ggplot(gt, aes(x = comp_length, y = variant_num)) +
geom_point(size = 3) + ylim(0, 1) +
labs(x = "Length of complement", y = "Choice of construction") +
scale_y_continuous(breaks = c(0, 1), labels = c("gonna", "going to")) +
theme_minimal(base_size = 20) + theme(aspect.ratio = 1)Logistic regression analysis
Methods of Corpus Linguistics (class 6)
Outline
- Introduction
- Probabilities, odds and logit
- Evaluation
- Interactions
- Random effects
Introduction
Set up code
Introduction
gonna dataset
Code
| variant | comp_length | register | variant_num | source | fit1 | fit2 |
|---|---|---|---|---|---|---|
| gonna | 6.72 | formal | 0 | q | 0.008 | 0.050 |
| gonna | 5.06 | informal | 0 | x | 0.348 | 0.137 |
| gonna | 5.28 | informal | 0 | z | 0.233 | 0.081 |
| gonna | 4.84 | informal | 0 | i | 0.480 | 0.221 |
| gonna | 5.74 | informal | 0 | m | 0.088 | 0.026 |
| gonna | 3.68 | informal | 0 | f | 0.946 | 0.861 |
| gonna | 5.15 | informal | 0 | j | 0.297 | 0.111 |
| gonna | 6.22 | informal | 0 | p | 0.027 | 0.007 |
| gonna | 6.60 | informal | 0 | p | 0.011 | 0.003 |
| gonna | 6.19 | formal | 0 | c | 0.029 | 0.174 |
| variant | comp_length | register | variant_num | source | fit1 | fit2 |
|---|---|---|---|---|---|---|
| going_to | 2.75 | formal | 1 | g | 0.995 | 0.999 |
| going_to | 4.22 | formal | 1 | r | 0.817 | 0.975 |
| going_to | 4.64 | formal | 1 | x | 0.608 | 0.929 |
| going_to | 4.77 | formal | 1 | q | 0.526 | 0.901 |
| going_to | 4.07 | informal | 1 | f | 0.869 | 0.691 |
| going_to | 1.78 | formal | 1 | y | 1.000 | 1.000 |
| going_to | 5.25 | formal | 1 | p | 0.251 | 0.723 |
| going_to | 4.29 | informal | 1 | g | 0.789 | 0.551 |
| going_to | 1.71 | informal | 1 | h | 1.000 | 0.999 |
| going_to | 1.00 | formal | 1 | c | 1.000 | 1.000 |
Introduction
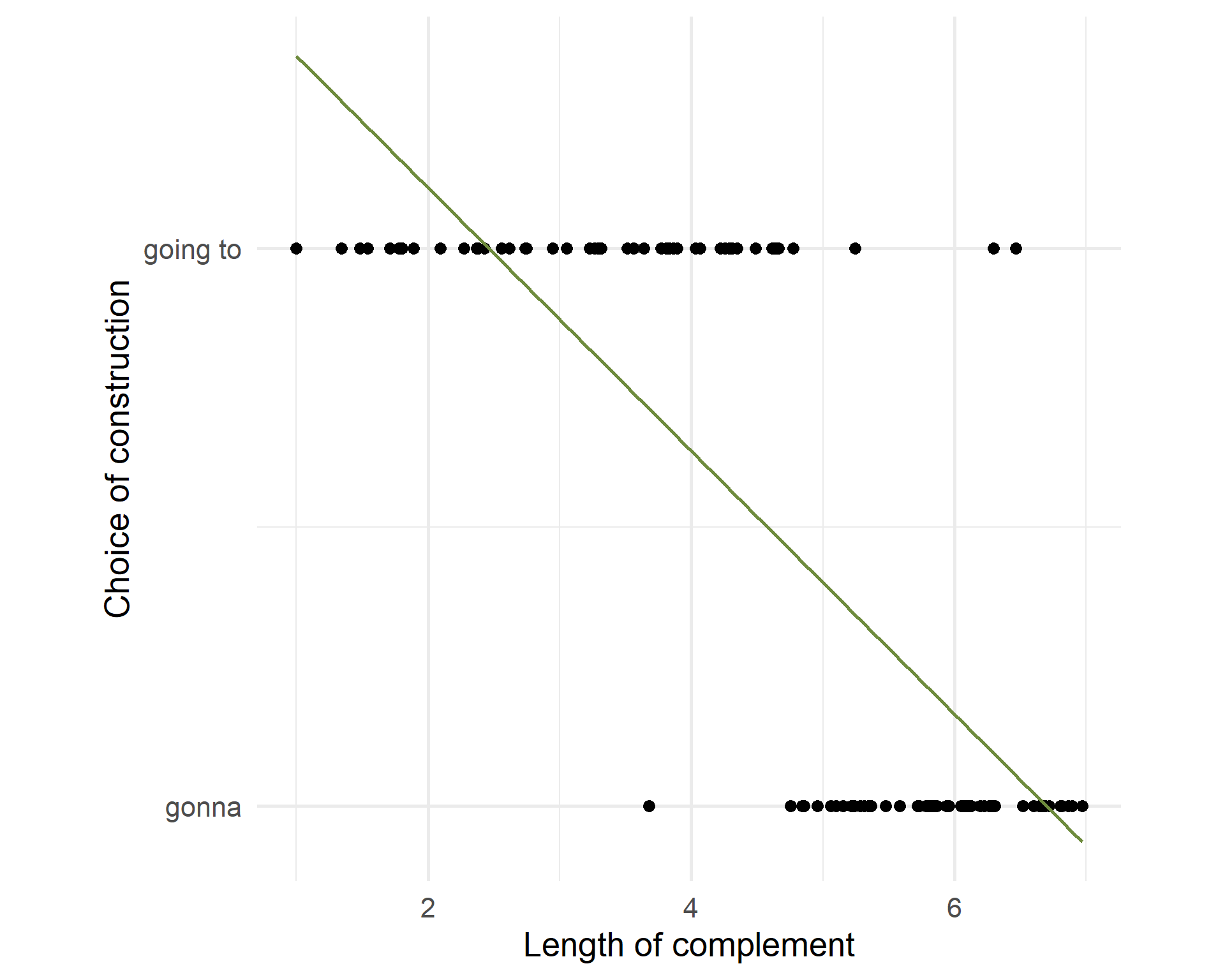
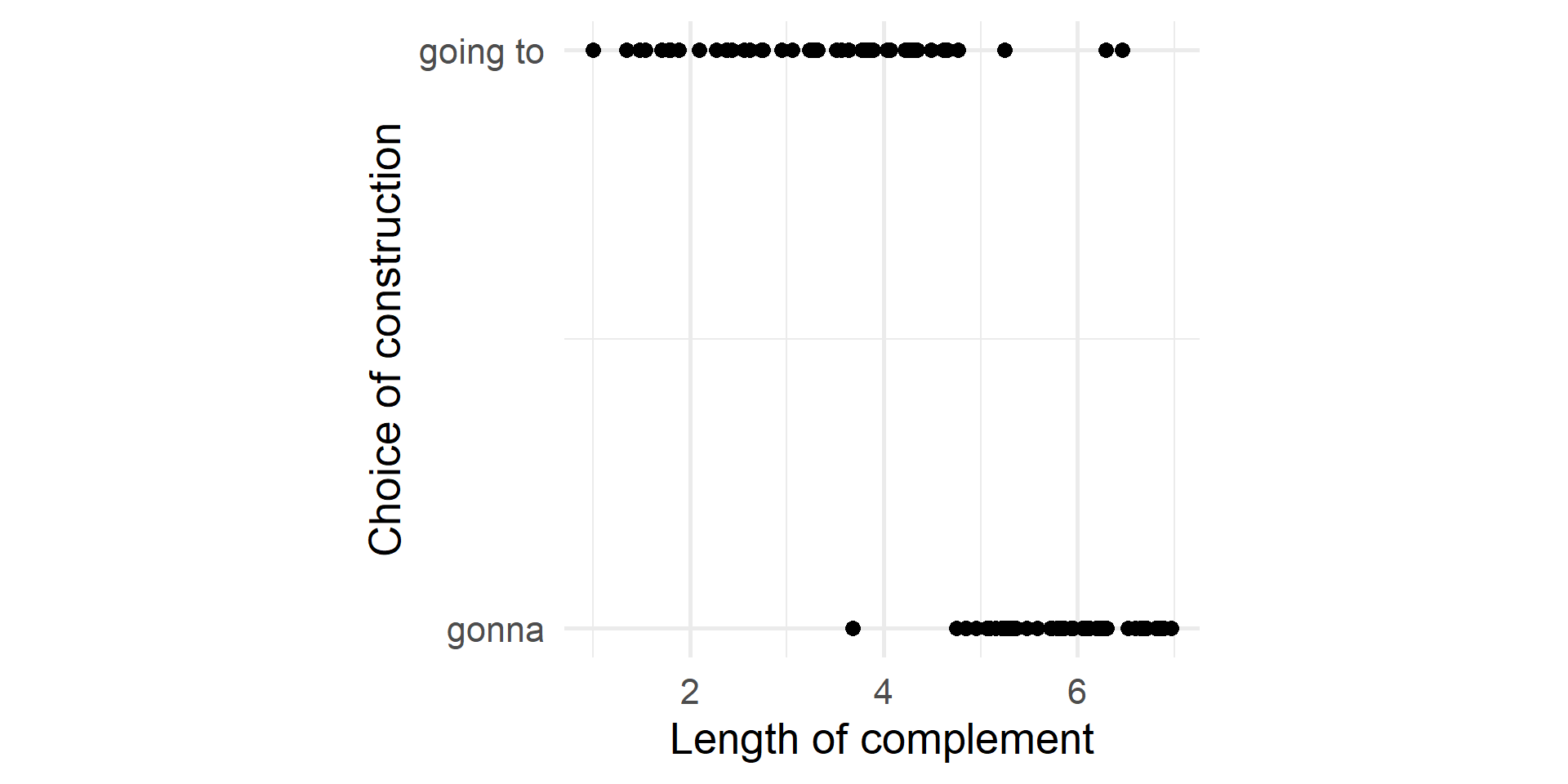
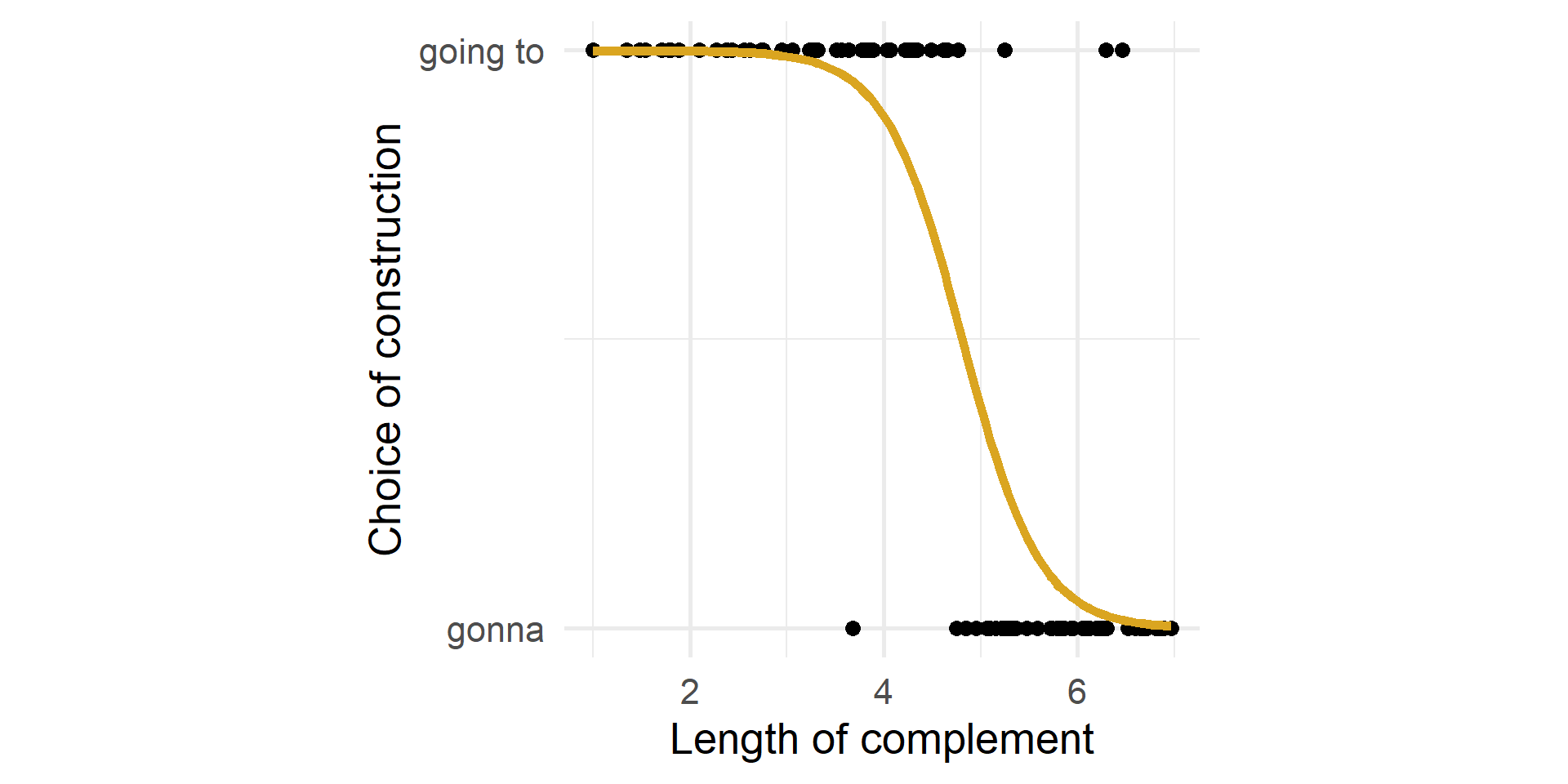
Scatterplot
What is the relationship between the length of the complement and the choice of going to/gonna?
Introduction
Linear fit
The relationship is not linear.
The residuals are not normally distributed.
Not all values are possible (probabilities go between 0 and 1).
Introduction
Logistic fit
The fit has an S shape.
Introduction
Model
Call:
glm(formula = variant ~ comp_length + register, family = binomial(logit),
data = gt)
Deviance Residuals:
Min 1Q Median 3Q Max
-1.9876 -0.2053 -0.0209 0.1067 2.1787
Coefficients:
Estimate Std. Error z value Pr(>|z|)
(Intercept) 11.582 2.899 4.00 6.5e-05 ***
comp_length -2.651 0.613 -4.32 1.5e-05 ***
registerformal 3.284 1.068 3.08 0.0021 **
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
(Dispersion parameter for binomial family taken to be 1)
Null deviance: 138.63 on 99 degrees of freedom
Residual deviance: 34.02 on 97 degrees of freedom
AIC: 40.02
Number of Fisher Scoring iterations: 7Introduction
Interpretation
Intercept
log odds of outcome (“going to”) when all predictors are at 0 (
comp_length= 0).odds =
exp(11.582)= 107138prob = odds/(odds+1) \(\approx\) 1
Coefficients
log odds ratios: positive increases chances of “going to”, negative of “gonna”.
odds ratio
of
comp_length=exp(-2.651)= 0.071of
register=exp(3.284)= 26.676
The odds of going to vs gonna in the formal register are 26.676 times higher than those in the informal register, other variables being controlled for.
Introduction
Probabilities, odds and logit
Probabilities
probs.
\(P\)
0 - 0.5 - 1
Number of successes divided by number of trials.
Probabilities, odds and logit
Odds
probs.
\(P\)
0 - 0.5 - 1
Number of successes divided by number of trials.
odds
\(\frac{P}{1-P}\)
0 - 1 - \(\infty\)
Probability of success divided by the probability of failure.
Undefined for \(P=1\).
Probabilities, odds and logit
Logit
probs.
\(P\)
0 - 0.5 - 1
Number of successes divided by number of trials.
odds
\(\frac{P}{1-P}\)
0 - 1 - \(\infty\)
Probability of success divided by the probability of failure.
Undefined for \(P=1\).
logit
\(\log\left(\frac{P}{1-P}\right)\)
\(-\infty\) - 0 - \(\infty\)
If positive, success is more likely; if negative failure is more likely.
Undefined for \(P=0\) and for \(P=1\).
Probabilities, odds and logit
Simulation
library(MASS) # to print fractions
probabilities <- c(1/c(7:2),
1-(1/c(3:7)))
probs <- tibble(
P = probabilities,
P_frac = as.character(fractions(P)),
odds = P/(1-P),
odds_frac = as.character(fractions(odds)),
logit = log(odds)
)
kbl(probs) %>% kable_paper(font_size = 22) %>%
row_spec(6, bold = TRUE)| P | P_frac | odds | odds_frac | logit |
|---|---|---|---|---|
| 0.143 | 1/7 | 0.167 | 1/6 | -1.792 |
| 0.167 | 1/6 | 0.200 | 1/5 | -1.609 |
| 0.200 | 1/5 | 0.250 | 1/4 | -1.386 |
| 0.250 | 1/4 | 0.333 | 1/3 | -1.099 |
| 0.333 | 1/3 | 0.500 | 1/2 | -0.693 |
| 0.500 | 1/2 | 1.000 | 1 | 0.000 |
| 0.667 | 2/3 | 2.000 | 2 | 0.693 |
| 0.750 | 3/4 | 3.000 | 3 | 1.099 |
| 0.800 | 4/5 | 4.000 | 4 | 1.386 |
| 0.833 | 5/6 | 5.000 | 5 | 1.609 |
| 0.857 | 6/7 | 6.000 | 6 | 1.792 |
Probabilities, odds and logit
Linear vs logistic relationships
Linear relation logit ~ x entails logistic curve p ~ x.
Probabilities, odds and logit
Evaluation
Model
Call:
glm(formula = variant ~ comp_length + register, family = binomial(logit),
data = gt)
Deviance Residuals:
Min 1Q Median 3Q Max
-1.9876 -0.2053 -0.0209 0.1067 2.1787
Coefficients:
Estimate Std. Error z value Pr(>|z|)
(Intercept) 11.582 2.899 4.00 6.5e-05 ***
comp_length -2.651 0.613 -4.32 1.5e-05 ***
registerformal 3.284 1.068 3.08 0.0021 **
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
(Dispersion parameter for binomial family taken to be 1)
Null deviance: 138.63 on 99 degrees of freedom
Residual deviance: 34.02 on 97 degrees of freedom
AIC: 40.02
Number of Fisher Scoring iterations: 7Evaluation
Deviance
- Null deviance
-
discrepancy between data and intercept-only model (like SST)
- Residual deviance
-
discrepancy between data and fitted model (like SSE)
Computation
\(-2 \log(L)\) with \(L\) the likelihood of encountering the data if the model is true.
- AIC (Akaike Information Criterion)
-
corrected residual deviance (to compare models with different N. of predictors) - the smaller the better
- Deviance residuals
-
contribution of each observation to the residual deviance.
Evaluation
Predicted probability
\[g(x) = \beta_0 + \beta_1x_1 + \beta_2x_2 + ...\]
Let’s predict the probability of going to when complement length is 4 and register is formal.
\[g(x) = 11.582 + -2.651\times 4 + 3.284 \times 1 = 4.26\]
odds = exp(4.26) = 70.802
\[P(x) = \frac{70.802}{70.802 + 1} = 0.986\]
Evaluation
Measures of predictive power
We take all pairs of observations in which the response variable is different (one of gonna, one of going to).
gt0is what the model predicted for the “gonna” element.gt1is what the model predicted for the “going to” element.
Code
| gt0 | gt1 |
|---|---|
| 0.011 | 0.999 |
| 0.221 | 1.000 |
| 0.009 | 0.999 |
| 0.050 | 1.000 |
| 0.021 | 0.895 |
| 0.001 | 1.000 |
| 0.793 | 1.000 |
| 0.006 | 0.942 |
| 0.090 | 0.998 |
| 0.265 | 1.000 |
| 0.006 | 0.514 |
| 0.019 | 1.000 |
| 0.174 | 0.093 |
| 0.111 | 0.420 |
| 0.003 | 1.000 |
Evaluation
C, D, T
C= concordant pair = prediction is higher for “going to” 😁D= discordant pair = the prediction is higher for “gonna” ☹️T= tie = the prediction is the same for both 😐
Code
| gt0 | gt1 | C | D | T |
|---|---|---|---|---|
| 0.011 | 0.999 | TRUE | FALSE | FALSE |
| 0.221 | 1.000 | TRUE | FALSE | FALSE |
| 0.009 | 0.999 | TRUE | FALSE | FALSE |
| 0.050 | 1.000 | TRUE | FALSE | FALSE |
| 0.021 | 0.895 | TRUE | FALSE | FALSE |
| 0.001 | 1.000 | TRUE | FALSE | FALSE |
| 0.793 | 1.000 | TRUE | FALSE | FALSE |
| 0.006 | 0.942 | TRUE | FALSE | FALSE |
| 0.090 | 0.998 | TRUE | FALSE | FALSE |
| 0.265 | 1.000 | TRUE | FALSE | FALSE |
| 0.006 | 0.514 | TRUE | FALSE | FALSE |
| 0.019 | 1.000 | TRUE | FALSE | FALSE |
| 0.174 | 0.093 | FALSE | TRUE | FALSE |
| 0.111 | 0.420 | TRUE | FALSE | FALSE |
| 0.003 | 1.000 | TRUE | FALSE | FALSE |
Evaluation
C-value
Code
| C | D | T |
|---|---|---|
| 14 | 1 | 0 |
\[c = \frac{C+T/2}{C+D+T} = \frac{14+0/2}{14+1+0} = 0.933\]
Linear vs logistic regression
| Linear regression | Logistic regression | |
|---|---|---|
| Response variable | Numerical | Categorical |
| Relationship between predictor estimate and response | Linear | Logistic1 |
| Fitting function | OSL (ordinary least squares) | MLE (maximum likelihood estimation) |
| Model comparison | F-test | AIC |
| Evaluation metric | \(R^2\) | \(C\) |
| Base R function | lm() |
glm() |
Evaluation
Interactions
What is an interaction?
When the effect of a predictor depends on the value of another predictor.
Formula: y ~ x1 + x2 + x1:x2 = y ~ x1*x2
Interactions
Examples
Possible interactions of complement length and register (is the text formal or informal?) on choice of gonna.
The effect of complement length is also positive/negative but stronger/weaker when texts are informal.
The effect of complement length is positive/negative when texts are informal, reversed when texts are formal.
Interactions
Model with interactions
Call:
glm(formula = variant ~ comp_length * register, family = binomial(logit),
data = gt)
Deviance Residuals:
Min 1Q Median 3Q Max
-2.5689 -0.0792 -0.0030 0.1584 1.8033
Coefficients:
Estimate Std. Error z value Pr(>|z|)
(Intercept) 18.78 7.10 2.64 0.0082 **
comp_length -4.22 1.55 -2.72 0.0065 **
registerformal -8.39 7.89 -1.06 0.2878
comp_length:registerformal 2.39 1.67 1.43 0.1529
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
(Dispersion parameter for binomial family taken to be 1)
Null deviance: 138.629 on 99 degrees of freedom
Residual deviance: 31.103 on 96 degrees of freedom
AIC: 39.1
Number of Fisher Scoring iterations: 8Interpretation
Intercept is interpreted in the same way.
comp_lengthis the effect of complement length in informal register.registerformalis the effect of the formal register at 0 complement length.comp_length:registerformalis the difference in the effect of complement length in formal register, compared to informal register.
Interactions
Visualization
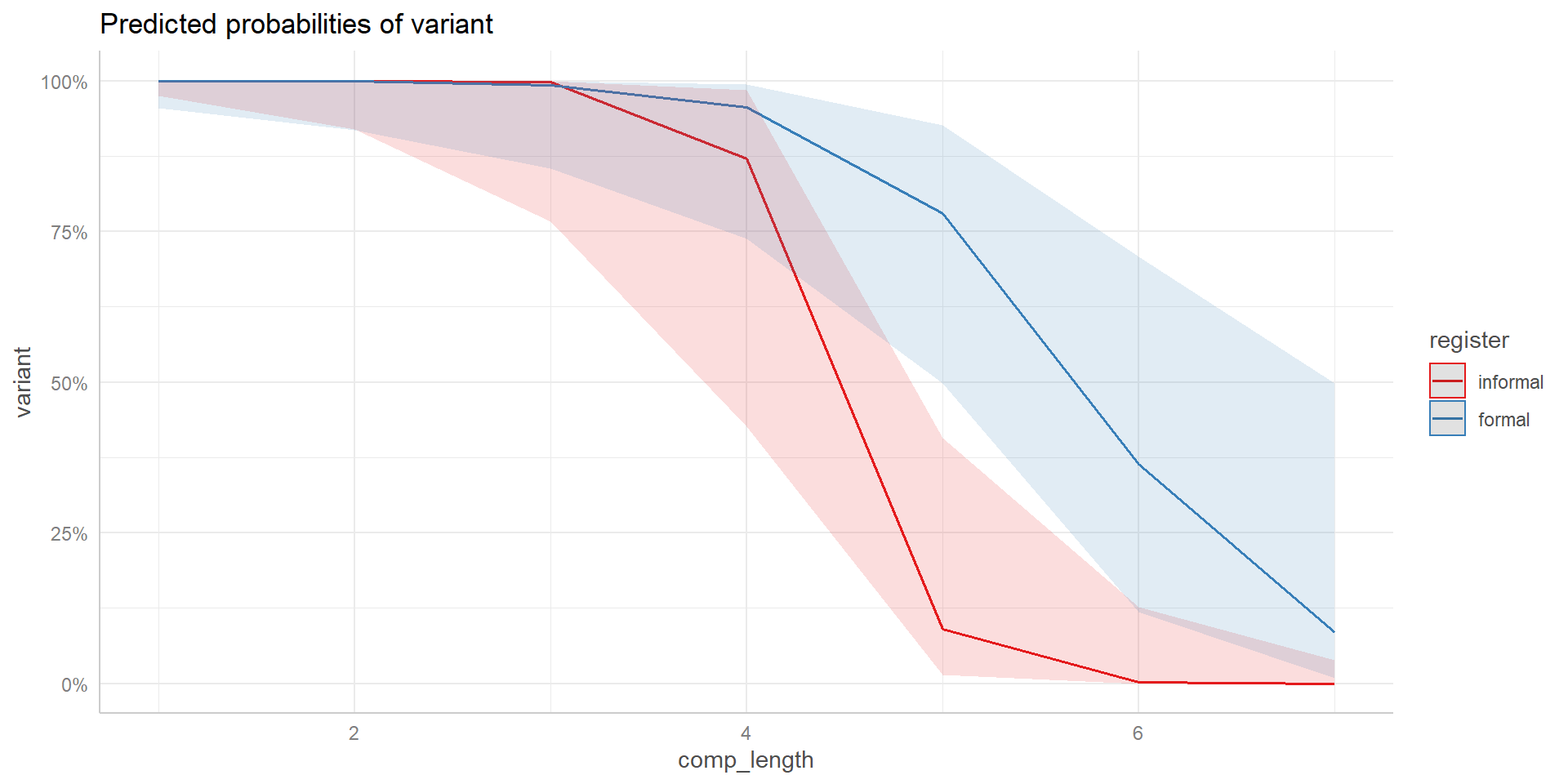
Interactions
Random effects
Fixed vs random effects
Unlike fixed effects (what we have dealt with up to now), random effects:
May not be replicable in a different dataset.
Involve a sample of the possible values in the population.
Examples: speaker, lexical effects, topic… → often operationalized as filename.
Random effects
Random intercept
The intercept can vary.
Different individuals prefer gonna over going to regardless of complement length.
Certain children always produce longer utterances than others of the same age.
Nested random intercept
E.g. random intercept for filename and random intercept for register; each filename occurs in only one register.
Random effects
Random slope
The slope of a fixed effect can vary.
A given predictor can have different effects in different texts.
Complement length is more influential in the choice of gonna for some individuals than for others.
For certain children, utterance length increases with age at a higher pace than of others.
Random effects
How to
lme4::glmer()instead ofglm();lme4::lmer()instead oflm().- The
controlargument helps us deal with convergence issues.
- The
Extension of the R formula:
(1 | filename)forfilenameas random effect.(age | child)forchildas a random slope forage.
Random effects
Model with random effects
m4 <- lme4::glmer(variant ~ comp_length*register + (1 | source), data = gt, family = binomial)
summary(m4)Generalized linear mixed model fit by maximum likelihood (Laplace
Approximation) [glmerMod]
Family: binomial ( logit )
Formula: variant ~ comp_length * register + (1 | source)
Data: gt
AIC BIC logLik deviance df.resid
40.7 53.7 -15.3 30.7 95
Scaled residuals:
Min 1Q Median 3Q Max
-4.041 -0.031 -0.001 0.063 1.343
Random effects:
Groups Name Variance Std.Dev.
source (Intercept) 1.6 1.26
Number of obs: 100, groups: source, 26
Fixed effects:
Estimate Std. Error z value Pr(>|z|)
(Intercept) 23.19 11.72 1.98 0.048 *
comp_length -5.18 2.56 -2.03 0.043 *
registerformal -10.30 9.77 -1.05 0.292
comp_length:registerformal 2.94 2.18 1.35 0.177
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Correlation of Fixed Effects:
(Intr) cmp_ln rgstrf
comp_length -0.997
registrfrml -0.829 0.827
cmp_lngth:r 0.890 -0.894 -0.985Random effects
Compare models
# Comparison of Model Performance Indices
Name | Model | AIC (weights)
-------------------------------
m1 | glm | 51.6 (<.001)
m2 | glm | 40.0 (0.303)
m3 | glm | 39.1 (0.479)
m4 | glmerMod | 40.7 (0.217)Random effects