Reading and exploring a corpus
Methods of Corpus Linguistics (class 2)
Outline
- Initialize project
- Add corpus
- Explore corpus
- Describe corpus
Initialize project
Steps
- Create an R project, check on the “git repository” checkbox and commit changes
- Alternatively, run
usethis::use_git()
- Add remote on the terminal following the instructions on GitHub (see next slide).
In the future
For future projects, the workflow may be different; check Happy Git With R for a guide.
Initialize project
Connect repository to GitHub
This only needs to be done once at the beginning.
Note
<url> is the url of your repository.
Initialize project
Notes
If you make changes on the remote, use
git pullbefore making changes in the local repo.Avoid .RData with
Tools > Global Options > General > Workspace/HistoryYou can work with Git(Hub) on the Git tab of RStudio or on the Git Bash Terminal
Initialize project
Add corpus
Git branch
Optionally, you can start “new work” on a new branch and then merge it to main.
Tip
I will only look at what you push to the main branch.
Add corpus
Add a folder with a corpus
Download the corpora from Toledo (mcl.zip file with various corpora) and copy/move the brown folder to your project. There are different options.
To be accessed with here::here("brown").
project
|_brown
|_project.Rproj
|_.gitignoreTo be accessed with here::here("corpus", "brown").
project
|_corpus
| \_brown
|_project.Rproj
|_.gitignoreTo be accessed with here::here("data", "corpus", "brown").
project
|_data
| \_corpus
| \_brown
|_project.Rproj
|_.gitignoreAdd corpus
.gitignore
We don’t want to track the corpus on git (because of size and licenses).
Open
.gitignore.Add a line for the folder to ignore, e.g.
/brown/.
(Check git status!)
Add corpus
Explore corpus
Create new R script
Easier to run the code again and to share it.
Load packages first.
Do not change your working directory; use
here()or reliable relative paths.
Tip
You can add comments to explain what you did and even hierarchical sections!
Explore corpus
Start of the script
Explore corpus
Inspect objects
In the console:
path_to_corpus=print(path_to_corpus)print(brown_fnames, hide_path = path_to_corpus)explore(brown_fnames)drop_re(brown_fnames, "/c[a-z]")
Explore corpus
Create a frequency list
Create it on the script, inspect in the console (or from the script).
Tip
Check out the “freqlists” tutorial of {mclmtutorials} (learnr::run_tutorial("freqlists", "mclmtutorials")) to learn why we need the re_token_splitter argument.
Explore corpus
Explore the frequency list
Frequency list (types in list: 63517, tokens in list: 1162192)
rank type abs_freq nrm_freq
---- ------ -------- --------
1 the/at 69013 593.818
2 ,/, 58153 500.373
3 ./. 48812 419.999
...[1] 1162192[1] 63517Tip
Check out the documentation
Explore corpus
Plot frequencies I
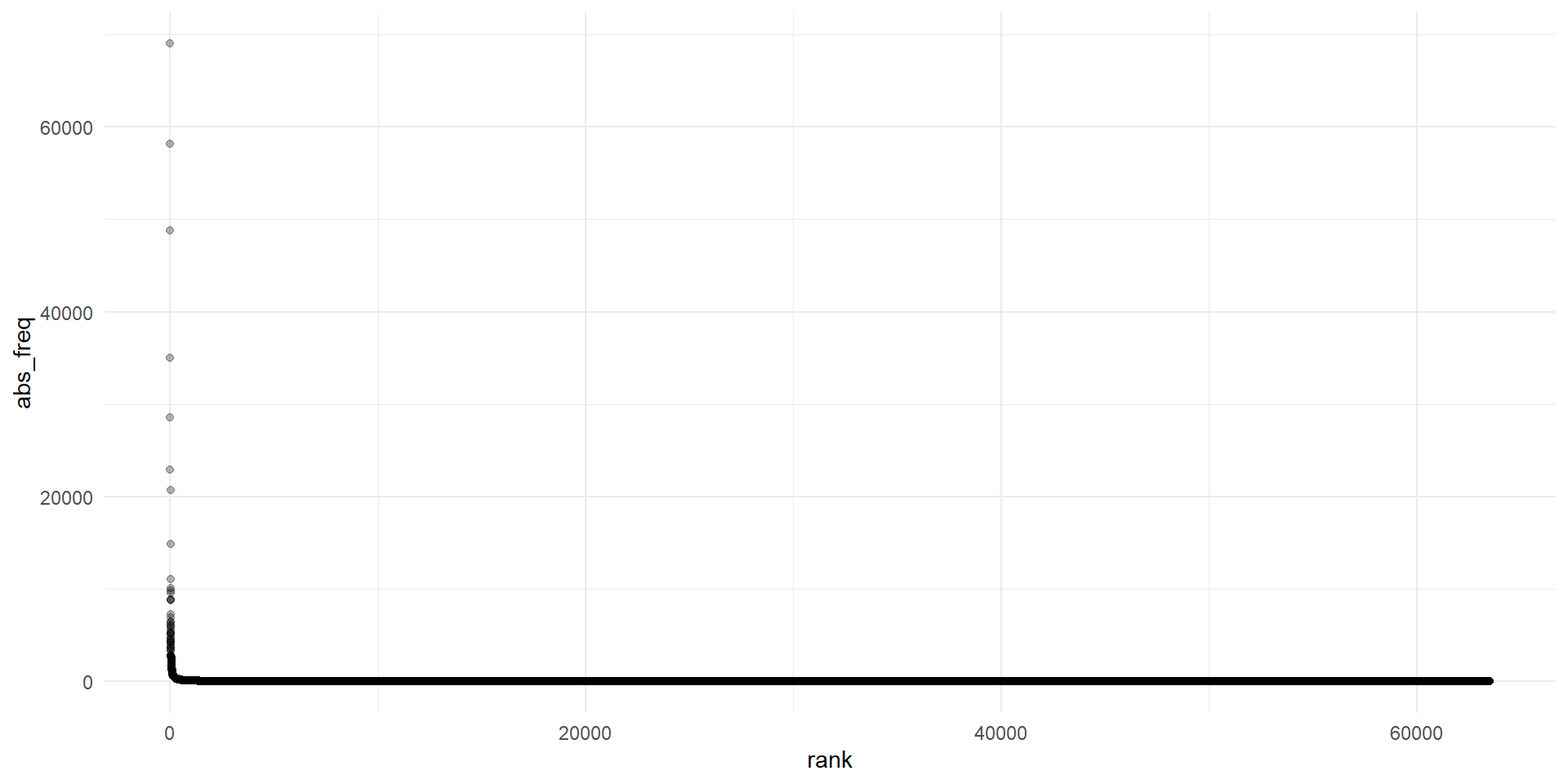
Explore corpus
Plot frequencies II
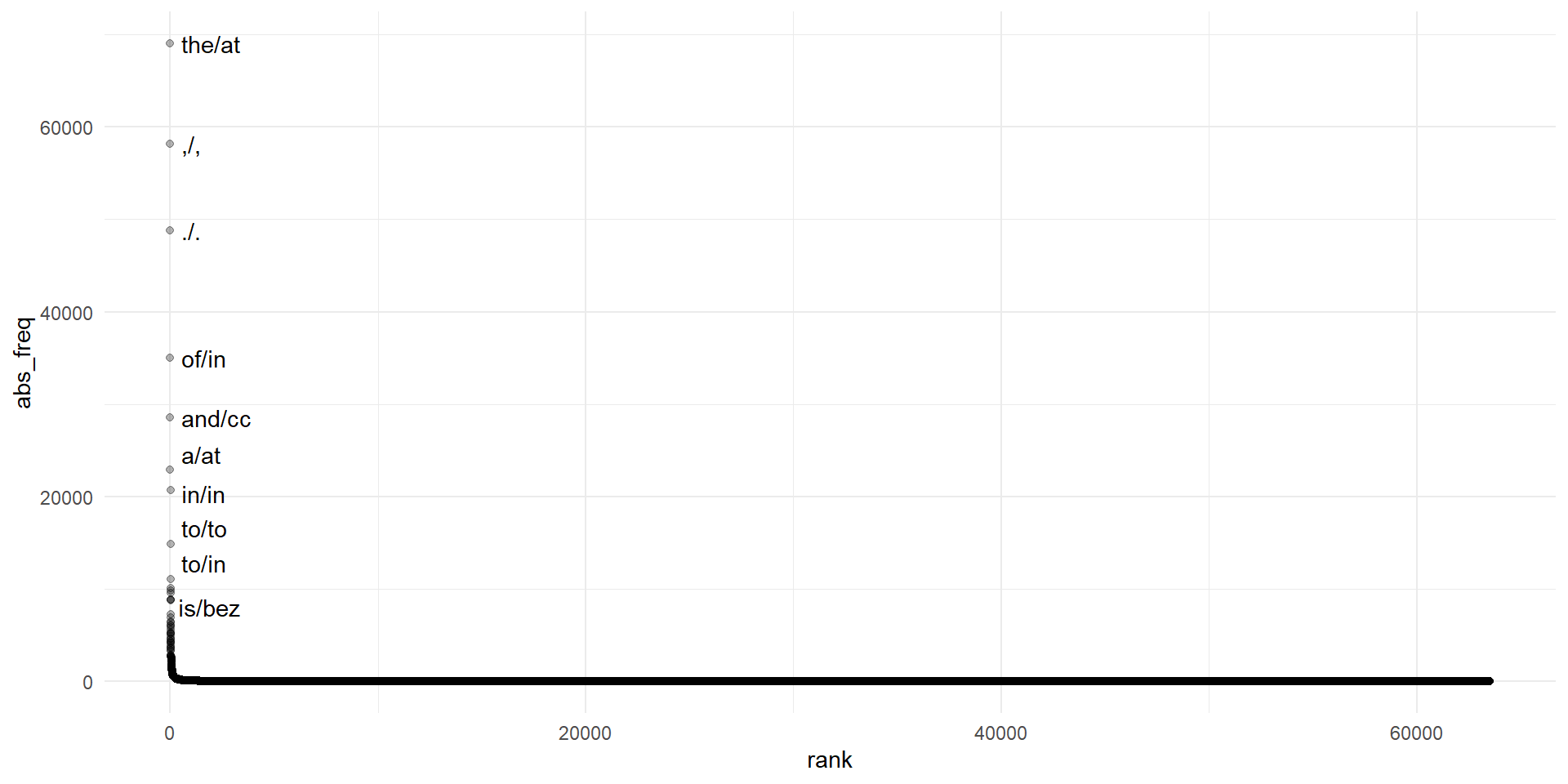
Explore corpus
Plot frequencies III
Explore corpus
Plot frequencies III
Describe corpus
Read the documentation
It might be in a README file, online, as a paper…
What time period(s) is/are covered?
What genre(s)? Language varieties?
Written? Transcripts of oral texts?
Is it a monitor corpus?
Licenses
Check also the permissions you have as user of the corpus.
Describe corpus
Quarto document
Create basic Quarto document
Set meta data on the YAML choosing output
Optional: render to check it’s working
Remove current text and write your own
Describe corpus
Code in a Quarto document
Inline code surrounded by backticks and starting with “r”.
Code chunks: to run arbitrary code, create tables and plots, print glosses with {glossr}.
Read external scripts with
source()or with thecodeorfilechunk options.
Describe corpus
Inline code
Markdown
The Brown corpus used in this project has `r prettyNum(n_tokens(flist))` tokens
and `r n_types(flist)` types,
giving us a type-token ratio of `r round(n_types(flist)/n_tokens(flist), 2)`.Output
The Brown corpus used in this project has 1162192 tokens and 63517 types, giving us a type-token ratio of 0.05.
Describe corpus
Code chunks
Frequency list (types in list: 5, tokens in list: 239548)
<total number of tokens: 1162192>
rank orig_rank type abs_freq nrm_freq
---- --------- ------ -------- --------
1 1 the/at 69013 593.818
2 2 ,/, 58153 500.373
3 3 ./. 48812 419.999
4 4 of/in 35028 301.396
5 5 and/cc 28542 245.588Describe corpus
Tables with knitr::kable()
| rank | orig_rank | type | abs_freq | nrm_freq |
|---|---|---|---|---|
| 1 | 1 | the/at | 69013 | 593.8175 |
| 2 | 2 | ,/, | 58153 | 500.3734 |
| 3 | 3 | ./. | 48812 | 419.9994 |
| 4 | 4 | of/in | 35028 | 301.3960 |
| 5 | 5 | and/cc | 28542 | 245.5876 |
Describe corpus
Tables with kableExtra::kbl()
| rank | orig_rank | type | abs_freq | nrm_freq |
|---|---|---|---|---|
| 1 | 1 | the/at | 69013 | 593.8175 |
| 2 | 2 | ,/, | 58153 | 500.3734 |
| 3 | 3 | ./. | 48812 | 419.9994 |
| 4 | 4 | of/in | 35028 | 301.3960 |
| 5 | 5 | and/cc | 28542 | 245.5876 |
Describe corpus
Import code
In both cases, you might want to use the include: false chunk option to avoid printing neither the code itself or its output.
Describe corpus
Stage, commit, push
- Commit whenever you reach a “stage”.
- Push at most once a day.
Branches and remotes
The first time you try to push from a local branch you may get an error! Just follow the instructions, don’t panic :)
From a branch
If you were in your explore-corpus branch and want to bring changes to main
Describe corpus